Abstract
Introduction: Myelodysplastic syndromes (MDS) are a group of heterogeneous hematological malignancies with complex molecular backgrounds mainly affecting the elderly. The incorporation of prognostic mutations into conventional risk scores, for example, the revised International Prognostic Scoring System (IPSS-R), can improve prognostic power. Recently, Bernard et al. developed a molecular IPSS (IPSS-M) on the basis of hematological variables and 31 putative genes. The aim of our study was to compare the prognostic value of IPSS-M with the IPSS-R.
Methods: Our study consisted of 852 patients with de novo MDS from 8/2016-9/2021. IPSS-R and IPSS-M scores were calculated to define different prognostic subgroups in our cohort. A targeted gene sequencing of 141 genes was performed in 592 patients from 8/2016 to 3/2020 and a 267-gene panel for 260 patients from 4/2020-9/2021. The statistical predictive power of the prognostic models was assessed by the concordance index (C-index). A higher C-index indicates a better quality of discrimination.
Results: 550 (64.6%) male and 302 (35.4%) female individuals were included. The median age was 56 years (inter-quartile ranges [IQR]: 44, 64 years) with 63.4% of subjects ≤ 60 years old. Compared with the IPSS-M group, patients in our group were younger (median age: 56 vs. 73 years, P<0.001), had higher IPSS-R scores (P<0.001) and worse IPSS-R-defined cytogenetics (P<0.001). Also, the hemoglobin levels, absolute neutrophil counts (ANC), and platelet counts were all significantly lower in Chinese subjects (P-values from 0.02 to <0.001).
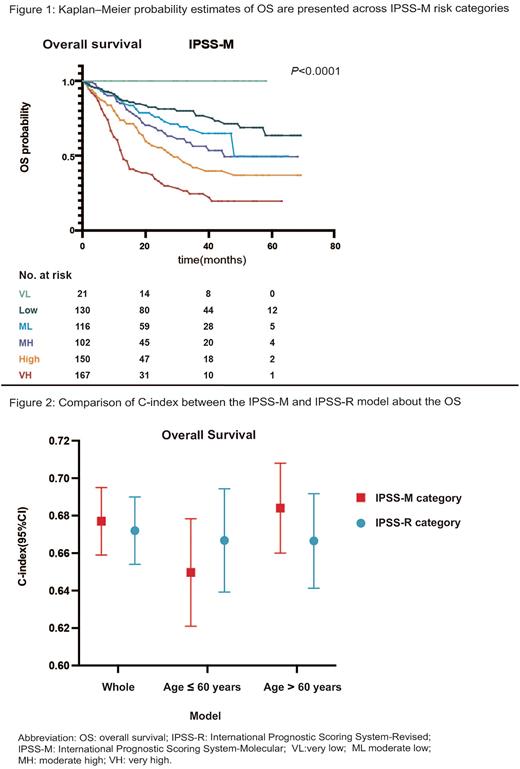
According to the IPSS-M, 21 patients (2.5%) were very low, 138 (16.2%) were low, 125 (14.7%) were moderate low, 113 (13.3%) were moderate high, 170 (20.0%) were high and 192 (22.5%) were very high, with a 3-year survival of 100%, 80%, 67.3%, 56.3%, 42%, 24.5% (P<0.0001, Figure 1). When analyzing the reclassification of patients from IPSS-R to IPSS-M, 247 (70.4%) were upstaged and 104 (29.6%) were downstaged. The median OS of intermediate IPSS-R patients reclassified as moderate, high and very high IPSS-M was not reached, 34 months and 13 months (P=0.014).
The C-index for the IPSS-R and IPSS-M were 0.67 and 0.68 in the whole group (Figure 2). Then we compared the discriminative power of the two models in younger group (age≤ 60 years) and older group (age> 60 years). IPSS-R had a higher C-index in younger group (0.67 vs. 0.65) while IPSS-M had a higher C-index in the elderly (0.69 vs. 0.66, Figure 2), suggesting that the predictive power of IPSS-M was more reliable than IPSS-R in older group.
To further analyze the differences between the younger and older ones, we compared the clinical, hematologic, cytogenetic and genetic data for both age groups. The older group was more likely to be male (72.1% vs. 60.2%, P<0.0001), had higher percentage of bone marrow blasts (P<0.0001) and worse IPSS-R karyotypes (P<0.0001). Considering the risk stratification, no significant difference was observed in IPSS-R categories between the younger and older ones (P=0.332). However, the elderly was enriched for very high IPSS-M category (27.9% vs. 19.4%, P=0.027).
The incidence and distribution of mutations among the two age groups were strikingly different. The average number of genes (2.0 vs. 1.6, P=0.003) and IPSS-M mutated genes (1.6 vs. 1.3, P=0.006) in the older ones was higher than the younger ones. The most frequent genes were ASXL1 in older MDS patients (21.5%) and U2AF1 in the younger ones (26.1%). The elderly had a higher incidence of RUNX1 (16.0% vs. 9.4%, P=0.004), TP53 (14.1% vs. 7.6%, P=0.002), TET2 (11.9% vs. 5.9%, P=0.002), SRSF2 (11.2% vs. 3.0%, P<0.0001), DNMT3A (10.9& vs. 6.7%, P=0.030), STAG2 (7.1% vs. 1.9%, P<0.0001), EZH2 (5.8% vs. 2.6%, P=0.019) and DDX41 (4.8% vs. 0.9%, P<0.0001), with U2AF1 more frequently occurring in younger ones (26.1% vs. 15.4%, P<0.0001).
Conclusion: The risk estimation differences between the IPSS-R and IPSS-M models in our cohort could be explained by study-specific factors, like unique ethnic groups and different population age. The IPSS-M could better stratify patients within the IPSS-R scoring system. With respect to an increased number of putative mutations and a different molecular profile in the elderly, IPSS-M model was more discriminative compared with IPSS-R in patients older than 60 years, but it had not been found to be more predictive in younger patients.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal